- About
- People
- Research
- Engagement
- Data
- Technical
- Search
Natural Language Processing
The pSCANNER Natural Language Processing (NLP) working group develops capacity to enable sharing and reuse of vast amounts of valuable health information derived from clinical texts of all pSCANNER participating institutes. Specifically, this involves creating generalizable and scalable NLP tools to extract phenotypes of the three use case conditions (i.e., Congestive Heart Failure, Obesity and Weight Management, and Kawasaki Disease) and store the results as a data warehouse of patient phenotypes in OMOP NLP schema proposed by the OHDSI NLP working group ready to support clinical studies. Currently, the working group provides the following NLP capacities for pSCANNER participating sites, as well as other CDRNs in PCORnet. Please contact pscanner@ucsd.edu for details.
The CLEAN cNLP Ensemble and Pipeline
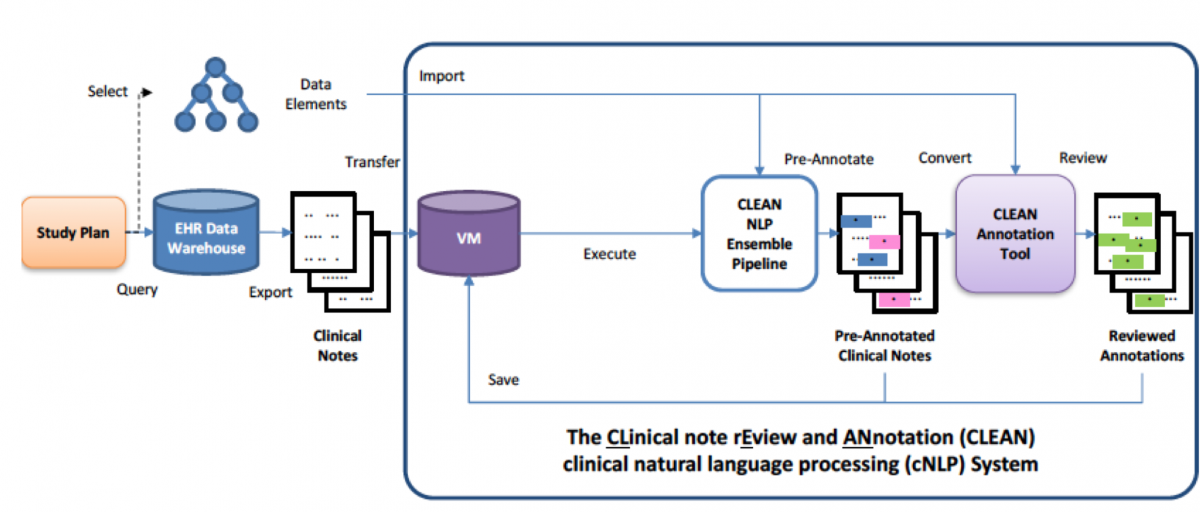
The CLEAN (CLinical note rEview and ANnotation) NLP ensemble pipeline is a secured, privacy-preserving platform for processing large clinical text corpora. CLEAN is able to assemble different NLP tools into an ensemble that synergizes strength of these tools to maximize the quality of the output. CLEAN also features an interactive annotation tool allowing secured visualization and review of the NLP, protected by the two-factor login and the virtual desktop. These features combine to provide high quality extraction from large corpora into a manageable time frame.
CLEAN provides the following NLP processing services:
- De-identification (de-ID )
- De-duplication
- Data element extraction
Please contact pscanner@ucsd.edu to use CLEAN. The CLEAN team will work with you to include your institution into an already approved IRB. Once it is approved, an account will be created for you to upload, process, and visualize your clinical text data securely.
The Leo Framework
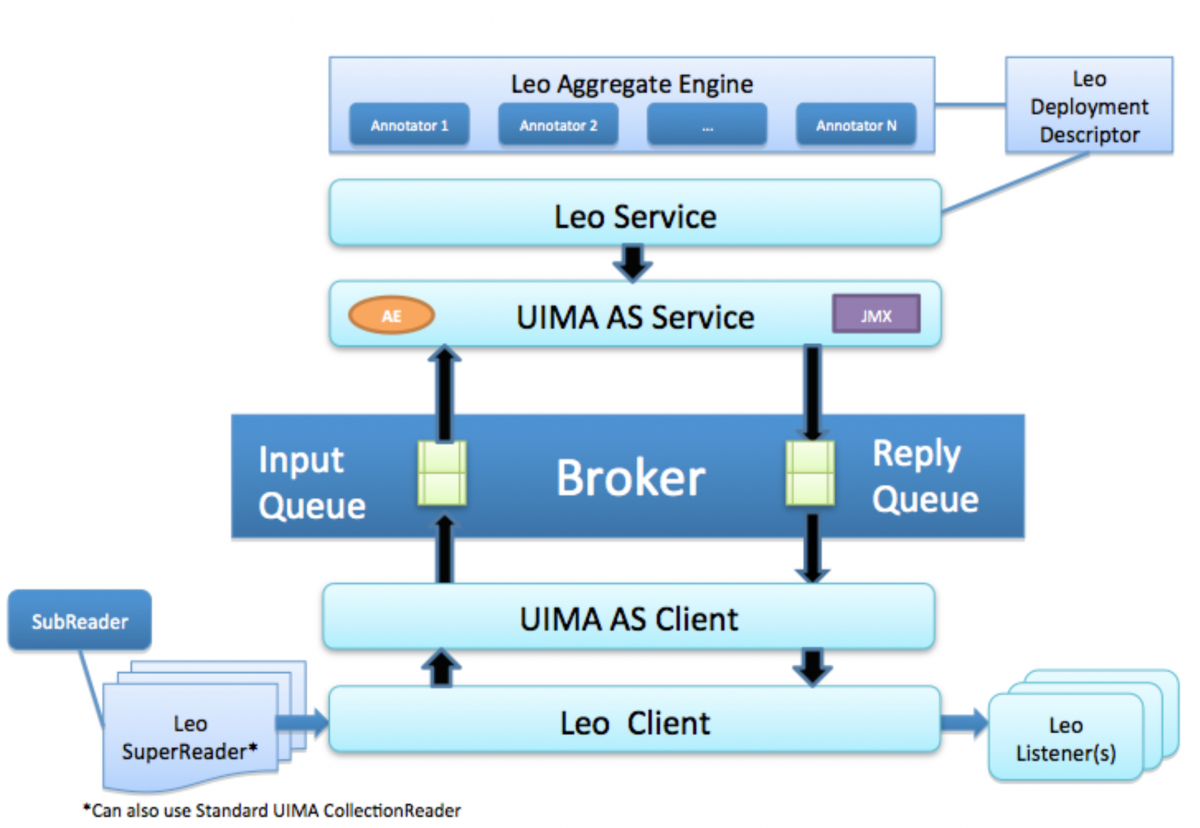
The VINCI-developed Natural Language Processing (NLP) infrastructure, is a set of services and libraries that facilitate the rapid creation and deployment of Apache UIMA-AS annotators focused on Natural Language Processing. The UIMA-AS underpinning allows Leo to manage the scale needed for real-time processing. It provides remote configuration tools for enabling automatic system optimization. With its developer utilities, functionality can be added and seamlessly integrated with existing NLP services. Leo enables users to programmatically generate primitive and aggregate UIMA analysis engine descriptors and deployment descriptors.
A detailed framework description and user guide can be found here:
http://department-of-veterans-affairs.github.io/Leo/index.html
If you use this system, please cite:
Cornia R, Patterson OV, Ginter T, Duvall SL. Rapid NLP Development with Leo. In: AMIA Annu Symp Proc.; 2014.
Vital Signs Extraction System
Assessment of vital signs is an essential part of surveillance of critically ill patients to detect condition changes and clinical deterioration. While most modern electronic medical records allow for vitals to be recorded in a structured format, the frequency and quality of what is electronically stored may differ from how these measures are actually recorded. We created a tool that extracts blood pressure, heart rate, temperature, respiratory rate, blood oxygen saturation, and pain level from nursing and other clinical notes recorded in the course of inpatient care to supplement structured vital sign data.
The system is available at: https://github.com/department-of-veterans-affairs/vitals/
If you use this system, please cite:
Patterson OV, Jones M, Yao Y, Viernes B, Alba PR, Iwashyna TJ, DuVall SL. Extraction of Vital Signs from Clinical Notes. Stud Health Technol Inform. 2015; 216:1035. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26262334
EchoExtractor
A natural language processing system using a dictionary lookup, rules, and patterns was developed to extract heart function measurements that are typically recorded in echocardiogram reports as measurement-value pairs. Curated semantic bootstrapping was used to create a custom dictionary that extends existing terminologies based on terms that appear in the medical record. A novel disambiguation method based on semantic constraints was created to identify and discard erroneous alternative definitions of the measurement terms. The system was built utilizing a scalable framework, making it available for processing large datasets.
The system is available at: https://github.com/department-of-veterans-affairs/echoextractor
EFEx
One of the most requested variables for research in heart disease is left ventricular ejection fraction (LVEF). LVEF is a measure of heart function and is expressed as the percentage of total blood volume pumped out of the heart with each beat. Only a small fraction of LVEF values are available as structured data in the VA electronic medical record system; the majority are recorded within notes and reports. To improve availability of LVEF data, VINCI has developed an NLP system to extract LVEF values from these clinical documents. The system was validated with precision of over 98% across different document types.
The system is available at: https://github.com/department-of-veterans-affairs/efex
CLAMP
CLAMP (Clinical Language Annotation, Modeling, and Processing) Toolkit is a comprehensive clinical NLP software that enables recognition and automatic encoding of clinical information in narrative patient reports. CLAMP components are built on proven methods in many clinical NLP challenges. CLAMP is customizable and allows users to choose from various choices of NLP and Machine Learning Components, annotate target documents, generate models, and process clinical notes.
To obtain an educational license to use CLAMP, please go to http://clamp.uth.edu/get-clamp.php

